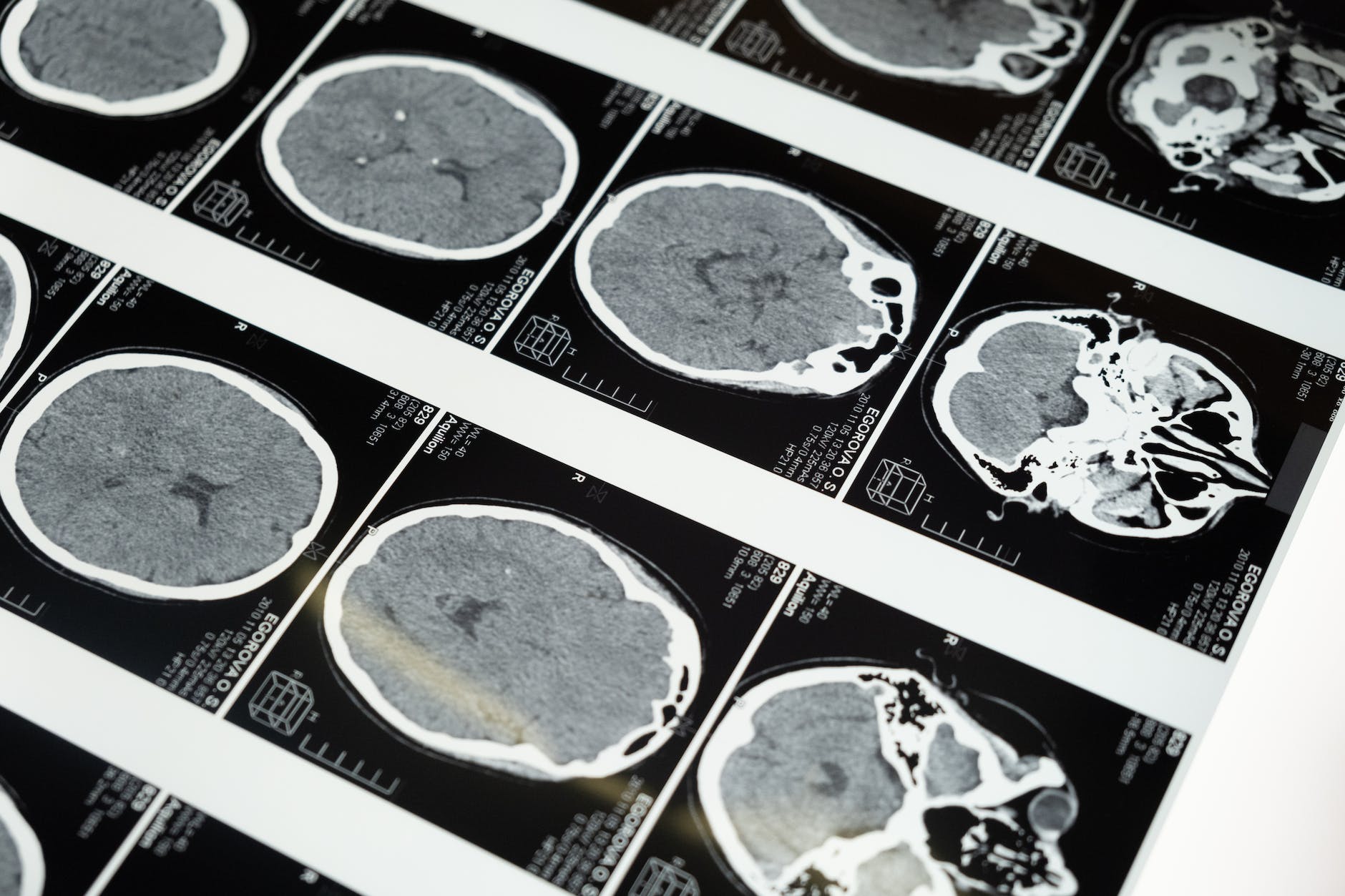
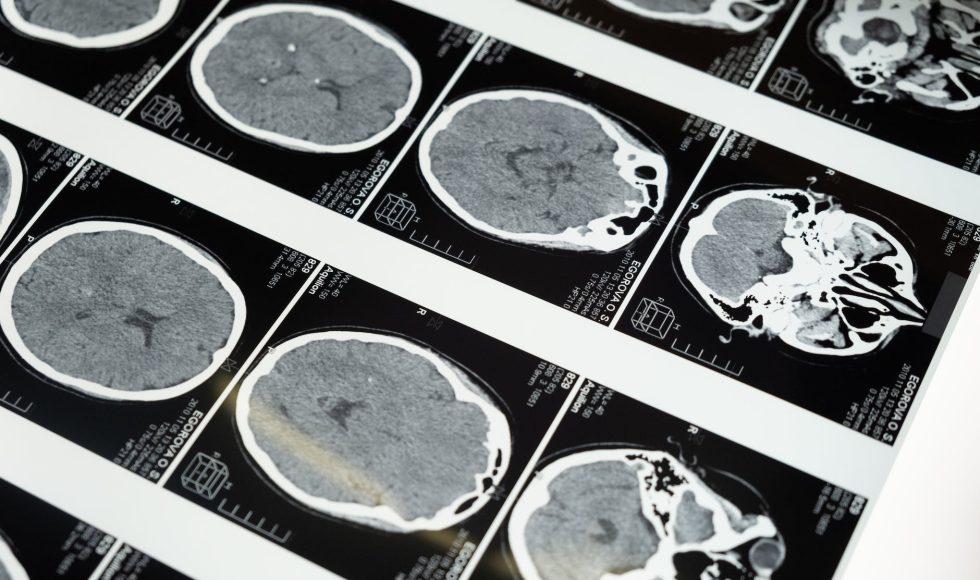
Franz-Josef Müller & Helene Kretzmer gave a presentation at the Nanopore Community Meeting 2021 about “Real-time cancer classification with nanopore sequencing.” Kretzmer defined real-time sequencing as “short time” sample processing from wet lab to bioinformatics. Muller explained that they have optimized a protocol for identification of brain cancers. They start with a rapid DNA extraction and the rapid sequencing kit (SQK-RAD004) with MIN106D flow cells. They also use customized software including Megalodon. Samples are taken directly from the operating room at the university hospital. Sample prep is immediately begun for the brain samples. The protocol can be performed by trained technicians. Within twenty minutes, the sample is ready for DNA quality control. The DNA is used for the Rapid Sequencing Kit using the MinION Mk1B (two!). In the video, they had two MinIONs. sequencing at the same time. The data come in within 43 minutes! Kretzmer explained that the sequences are processed in batches of reads with the help of Megalodon. They shared data of 500 CpGs within a couple of minutes. Within a couple of hours, the number of CpG identified over time flattens. The classification algorithm consists of a training step (of the model) and a run for classification. They calculate methylation kernels for each class. Then, these data are used to predict the tumor class based on the methylation rates. The team has devised a hierarchical system for classification of brain tumor classes. Kretzmer shared an animation of real-time classification. The slide had MethyLYZR listed as one of the software packages used. Over time, the set of tumors becomes smaller and more specific. The readout consists of three graphics: tumor classes and probabilities. The workflow is really efficient. I wonder how they purify DNA their DNA. Their video was really impressive: Muller called them clinical demonstrator videos to emphasize how the procedure can be used in the operating theatre.