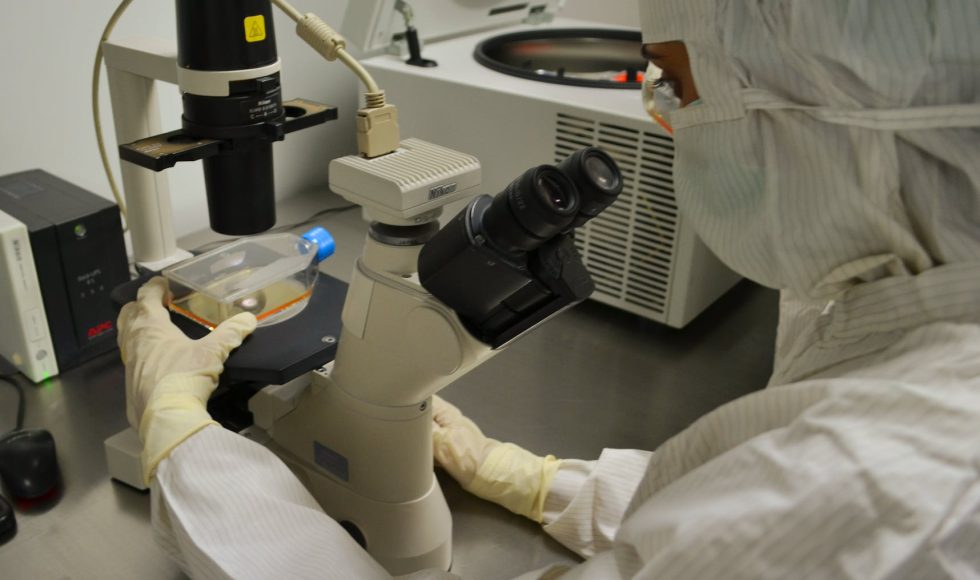
Rob James from the University of Warwick in the UK presented a session titled “From amplicons to metagenomes: long-read sequencing the environment” at London Calling 2019. James spoke about complex environmental matrices such as soil, sediment, and excreta that are often impure and biologically and chemically contaminated. Different extraction methods are used depending on the […]
Harika Urel, a Ph.D. student in Lara Urban’s lab at the Helmholtz Al Institute and Technical University of Munich in Germany, presented at London Calling 2023 on “Squiggle analysis for metagenomic viability inference.” I watched this session a couple of months ago and wanted to return to it in preparation for summer research and the […]
Marcela Aguilera Flores from Virginia Tech presented at the Nanopore Community Meeting 2019 on “Culture-free detection of boxwood blight to improve disease diagnosis and prevention.” Aguilera Flores spoke about the use of the MinION for metagenomics to detect the fungi that cause blight in the boxwood. According to Aguilera Flores, boxwood is an important ornamental […]
Eric Bortz is a professor at the University of Alaska Anchorage. Bortz presented at London Calling on “MinION Below Zero: sequencing microbes and marine mammals, birds, and the Alaskan coastal environment from low-quality samples, including whale snot”! What a title! I watched this session today to learn about the variety of samples they are sequencing. […]
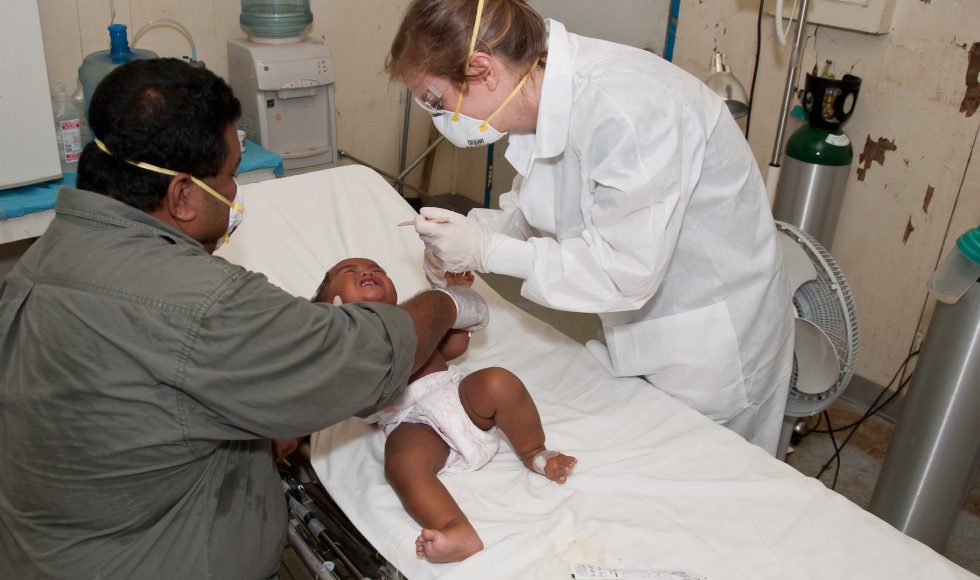
Hugh E. Olsen from the University of California at Santa Cruz spoke at the Nanopore Community Meeting 2021 about “Detecting SARS-CoV-2 and other pathogens in aerosols at wastewater treatment plants.” Just this past week, I was talking about this with a graduate student. Olsen said that “wastewater is a catalog of human pathogens.” Bioaerosols are […]
Weizhi Chen from Simcere Diagnostics presented a lightning talk at the Nanopore Community Meeting 2017 titled “Nanopore metagenomics sequencing for identification of both culture-positive and culture-negative pathogens.”Chen explained how they are using MinION and sequencing for infectious disease diagnostics. These labs are associated with labs in China. The first case report Chen shared was the […]
Teresa Street from the University of Oxford in the UK presented at the Nanopore Community Meeting 2019 on “Metagenomic nanopore sequencing for analysis of orthopedic device-related infection.” Street spoke about the impact of hip, knee, shoulder, elbow, and ankle replacements and the percentage of revisions due to the suspicion of infection. Further surgeries are often […]
Matthew Clark from The Genome Analysis Center presented at the Nanopore Community Meeting 2016 a session with the title “MinION metagenomics: from mock communities to clinical samples.” They took a mock community with twenty equimolar genomes. They were impressed with the results: 98%+ aligned, and there were long alignments. At the time, the group used […]
Mads Albertsen, Associate Professor at Aalborg University, presented at the Nanopore Community Meeting 2017 on “Genome-centric metagenomics in the long-read era.” Albertsen talked about how we live in a bacterial world. However, only a small fraction can be grown. Single-cell genomics is challenging. Metagenomics has the potential to separate genomes through binning. Now you can […]
Tim Walker from the Technical Services Team with Oxford Nanopore Technologies presented the Nanopore Learning Course – Metagenomics lesson I watched tonight. They spoke about metagenomic classification techniques to identify organisms from sequence data and annotate genes. For targeted metagenomics, ITS, 16S, or antibiotic resistance genes can be amplified and sequenced. Whole genome sequencing can […]