Tonight, I watched a shorter Nanopore Community Meeting Boston session. Anastasia McKinlay from Indiana University & HHMI presented on “Positional mapping of active versus silenced rRNA gene clusters within A. thaliana nucleolus organizer regions. McKinlay spoke about nucleolus organizer regions in Arabidopsis thaliana. These positions are difficult to understand with shorter reads. Long-read sequencing helped […]
Individual cell analysis is possible with Oxford Nanopore Technologies (ONT). Matt Parker from ONT spoke about “Individual cells matter – single-cell data analysis with EPI2ME.” EPI2ME cloud was launched, and cloud-based analyses are possible. I have been testing and mentioning the new version in the Portable Genome Sequencing course. Bryan Leland, a Bioinformatics Manager with […]
Matt Parker, the Director of Clinical Bioinformatics Software at Oxford Nanopore Technologies, facilitated the Nanopore Community Meeting Boston 2024 session on variant analysis with EPI2ME. The title of the presentation was “Ultra-rich human data – variant analysis with EPI2ME.” They shared the EPI2ME user interface updates that were recently released. There is a new element […]
Dmitrijs Rots from Erasmus MC in the Netherlands presented at the Nanopore Community Meeting in Boston (NCM Boston 2024) on “DNA methylation signature detection using ultra-rapid, long-read nanopore genome sequencing.” Rots mentioned that Erasmus MC is one of the largest hospitals in Europe! The team wanted to implement nanopore sequencing to test for methylation and […]
Tonight, I went into the lab to wash flow cells and try to reload the cDNA sequencing run. I may have caused more damaged… so I came back and watched the session “Individual cells matter – single-cell answers through nanopore sequencing.” Rachel Rubinstein, a Technical Product Manager at ONT, facilitated this showcase session. Rubinstein spoke […]
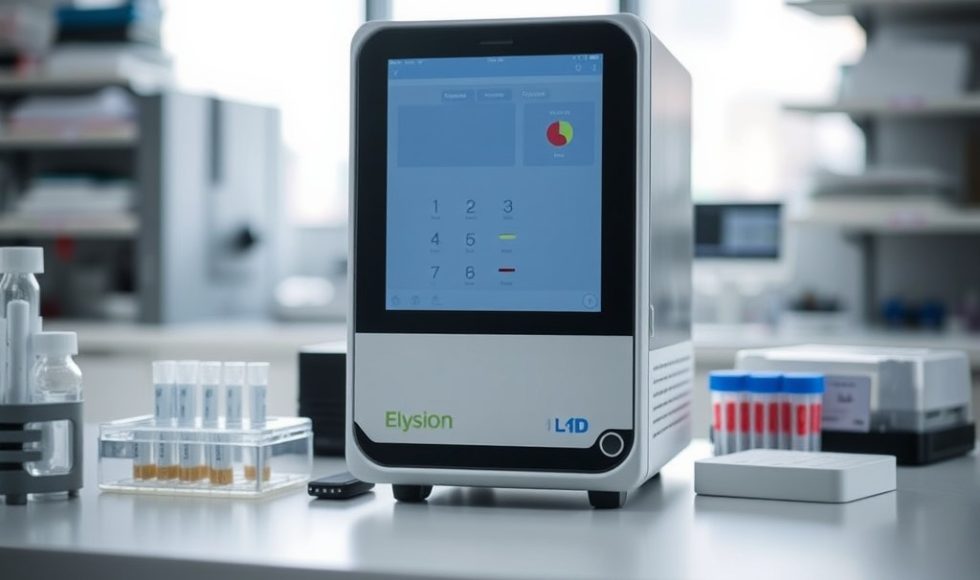
Jade Bartolo, the Associate Director for Automation at Oxford Nanopore Technology (ONT), introduced the ElysION. The five-minute recording is titled “Smarter genomics for streamlined answers – ElysION, sample-to-answer nanopore sequencing.” The ElysION has an integrated thermocycler and a MinION Mk1D or PromethION P2. The system can also shake and is compatible with tubes and plates. […]
Tonight, I watched the second half of a new Knowledge Exchange session focusing on “Sequencing and analysis of nanopore-only microbial isolates with the NO-MISS workflow.” Different extraction methods produced varying yields. Bead-beating and enzymatic lysis extractions affect read length and throughput, while fungal samples produce lower yields. Some potential issues include incomplete lysis and contaminants. Additional clean-up […]
We hosted a bioinformatics workshop with Oxford Nanopore Technologies specialists! It was fun to learn about EPI2ME updates. Tonight, I watched the beginning of a new Knowledge Exchange session focusing on “Sequencing and analysis of nanopore-only microbial isolates with the NO-MISS workflow.” The webinar began with a review of the applications related to microbial isolate […]
I continued watching the “Enhanced microbial profiling and metagenomics with nanopore sequencing” webinar. The second speaker was Steven Batinovic, a Field Application Scientist with Oxford Nanopore Technologies (ONT). The title of their session was “What you’re missing matters: delivering the future of microbial genomics with Oxford Nanopore.” Batinovic started by listing the microbiology and infectious […]
Tonight, I watched a Nanopore webinar by Tianyuan Zhang. The session’s title is “The Newest Oxford Nanopore R10.4.1 Full-length 16S rRNA Sequencing Enables the Accurate Resolution of Species-Level Microbial Community Profiling.” Zhang spoke about the resolution gains with full-length 16S sequencing, citing a Nature article from 2019. However, the error rate of ONT was previously […]