Tonight, I watched the London Calling 2019 session “Ultra-long reads and ultra-long duplications: deciphering the mysteries of the Bordetella pertussis genome.” Natalie Ring from the University of Bath in the UK was the presenter. They spoke about whooping cough and how the introduction of vaccination in the 1950s reduced the number of reported cases. In […]
Tonight, I watched Miten Jain’s London Calling 2019 presentation on “Generating high-quality reference human genomes using PromethION nanopore sequencing.” Jain is from the University of California, Santa Cruz. They have developed a framework to produce reference-quality human genomes in a week. Jain spoke about the need to sequence many human genomes that can serve as […]
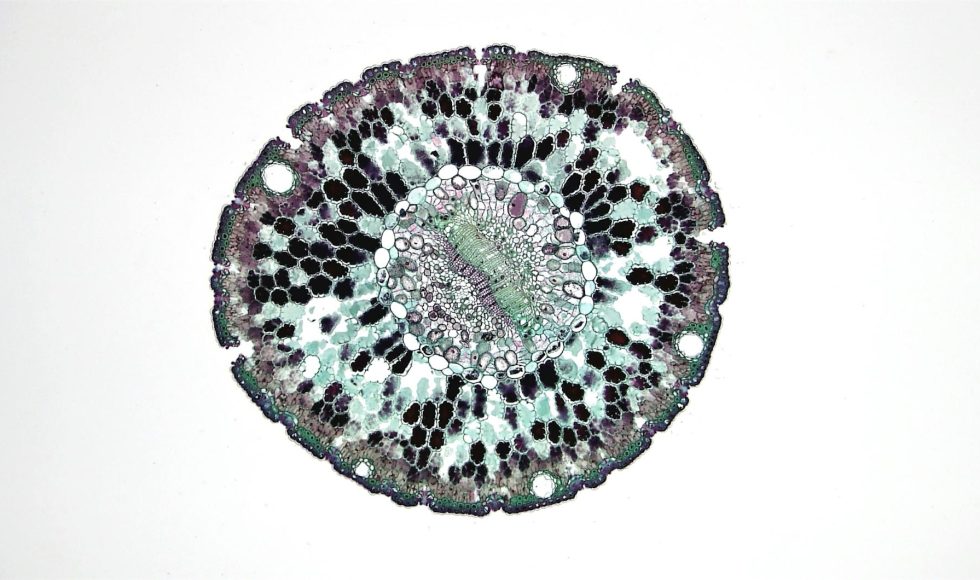
Vania Costa from Oxford Nanopore Technologies has recorded several Master Classes and sessions with ONT. I watched the London Calling 2019 session “The fever tree: extracting and preparing the DNA of Cinchona pubescens.” This plant produces quinine that is used for malaria and fever treatment. The team wanted to sequence the Cinchona genome to learn […]
Andrea Riposati from Dante Labs in the US presented at London Calling 2019 on “Human long-read whole genome sequencing: applications and results.” Riposati is the co-founder and CEO of Dante Labs. They presented a case of a patient who had suffered 45 years without a diagnosis. Dante Labs provided whole genome sequencing and pharmacogenomics lab. […]
Aaron Brooks from EMBL Germany presented at London Calling 2019 on “Measuring SCRaMbLEd transcription with direct RNA sequencing.” Direct RNA sequencing has interested me since we tried it last summer for bacterial transcriptomics. Brooks asked: “What if you could build a genome from scratch?” and “Where would you place the transcriptional units?” It is now […]
Emma Langan from the Earlham Institute in the UK spoke at London Calling 2019 on “Ship-Seq: nanopore sequencing of polar microbes onboard research vessels.” This was Langan’s Ph.D. project. Phytoplankton are photosynthetic organisms that live in water. They are responsible for carbon cycling and biogeochemical cycling. Langan is interested in diatoms, which are mainly found […]
Mike Clark from the University of Melbourne in Australia presented at London Calling 2019 on “Deep transcriptomic sampling with long-read single cell RNA sequencing.” Clark gave the first talk in the session and explained the power of expression profiles of single cells (scRNA-seq) to identify cell types and variations in gene expression. scRNA-seq can be […]
My Linh Thibodeau from the University of British Columbia in Canada presented at London Calling 2019 on “Resolution of germline hereditary cancer structural variants using nanopore sequencing.” They began talking about the Personalized OncoGenomics Program (POG), which is an initiative of BC Cancer. The study enrolls participants and conducts extensive genomic analyses. The team evaluated […]
Today was Aurelio’s fifth birthday! Tonight, I watched Timothy Gilpatrick’s London Calling 2019 session on “Targeted nanopore sequencing with Cas9 for studies of methylation structural variants and mutations.” Gilpatrick is a Johns Hopkins University. They are interested in specific loci and want to generate high coverage of those areas to examine structural variation. The enrichment […]
Pore-C is a method I hear about, yet I don’t fully understand the details. Eoghan Harrington from Oxford Nanopore Technologies (ONT) presented at London Calling 2019 a session titled “Pore-C: a method for genome-wide, multi-contact chromosome conformation capture.”Harrington is part of the applications team and focuses on genomic applications. They collaborate with various partners. The […]